As an introduction to working with the unPAK database this tutorial will walk you through the process of identifying plant lines of based on a certain phenotype of interest. To access the unPAK database click here (will open in new window).
STEP 1:
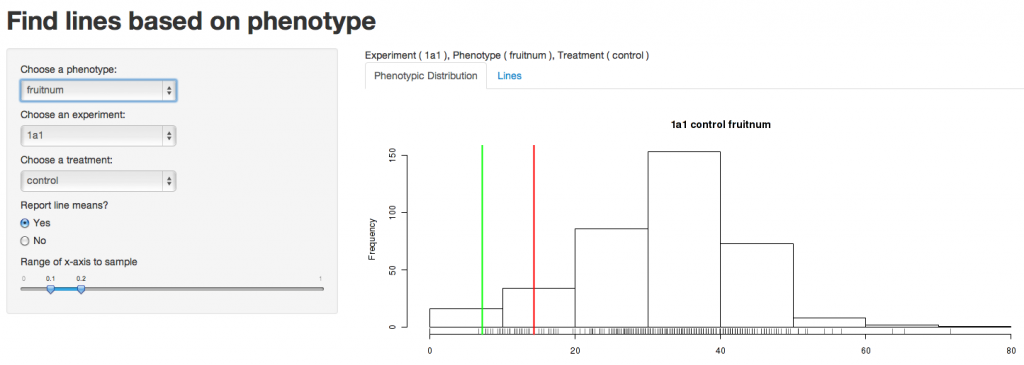
The above page is comprised of two parts, menus on the left that allow you to chose specific traits, experiments, treatments and trait value ranges of interest and a graph on the right that changes to reflect the options you have chose. For this example, start by choosing a phenotype, an experiment, and a treatment using the drop down menus. For this tutorial, we’ve chosen “fruitnum” (fruit number at time of harvest) as our trait, Experiment 1A1 as our experiment, and the control treatment as our treatment of interest.
STEP 2:
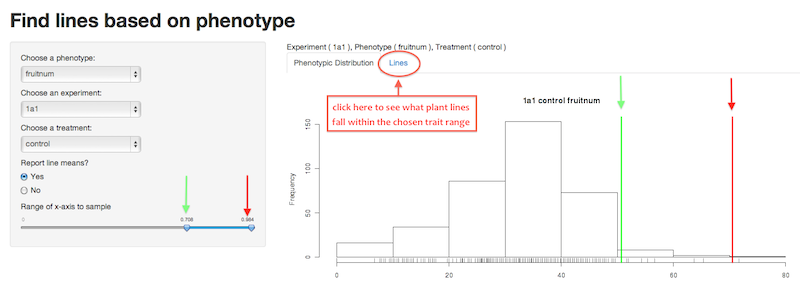
Now, to identify lines that fell within a certain range of trait values move the two blue arrowheads along the trait range slider in the left panel. As you move the arrowheads you will see the green bar (lower trait value cutoff) and red bar (upper trait value cutoff) move on the histogram.
Next, click on the link titled “Lines” above the graph to find out what Arabidopsis lines tested fall within the trait value range you selected.
STEP 3:
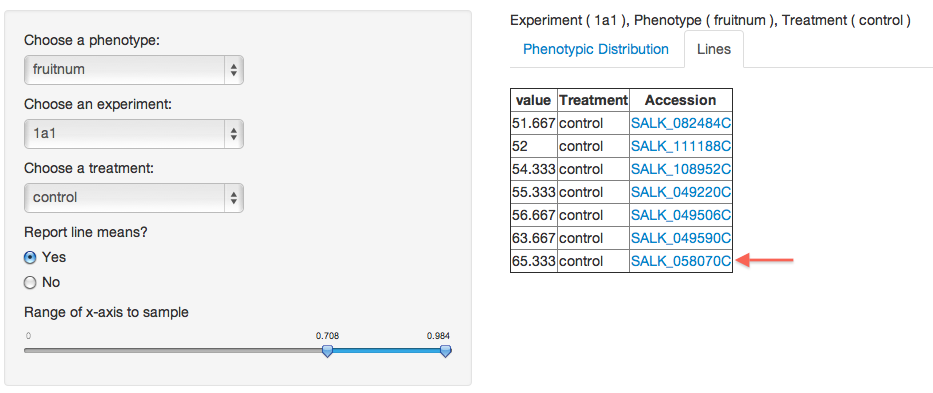
All of the line IDs listed under the “Accession” header in the table are links – clicking on them will bring you to a new page where you are able to view all of the phenotypes recorded for your plant line of interest. For the case of this tutorial, the figure above shows the results returned for plant lines used in Experiment 1A1 that on average produced the greatest number of fruits. We’ve chosen to select the mutant line SALK_058070C.
STEP 4: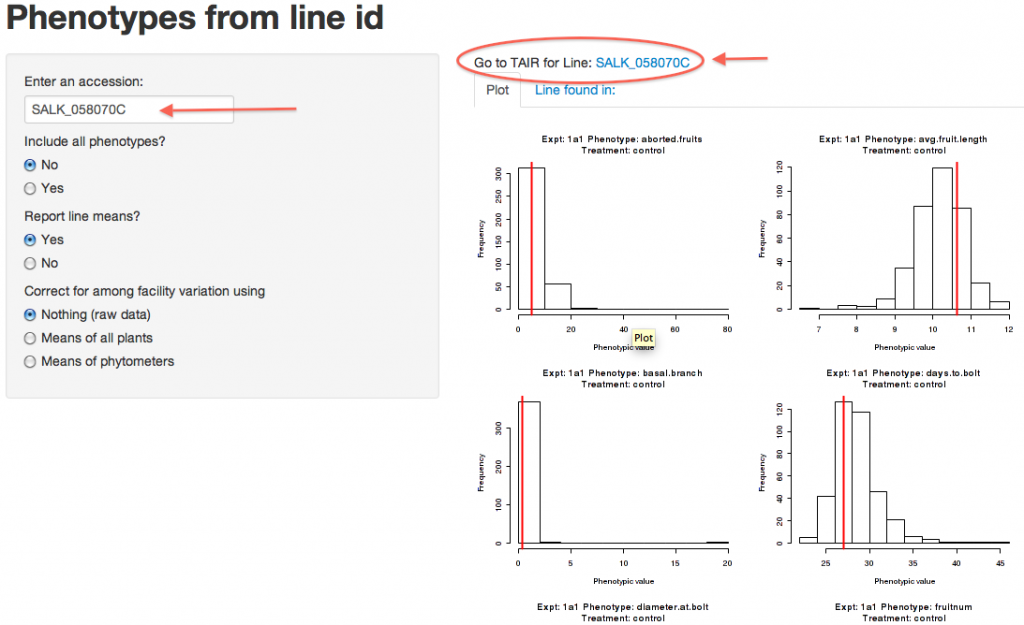
The “Phenotypes from line ID” interactive webpage allows you to view customized results for any plant line unPAK researchers have recorded data for. On the right are histograms of trait values of all plants separated by experiment and trait. The line of interest (in this case, SALK_058070C) is represented by a red line (the mean value of all replicates measured).
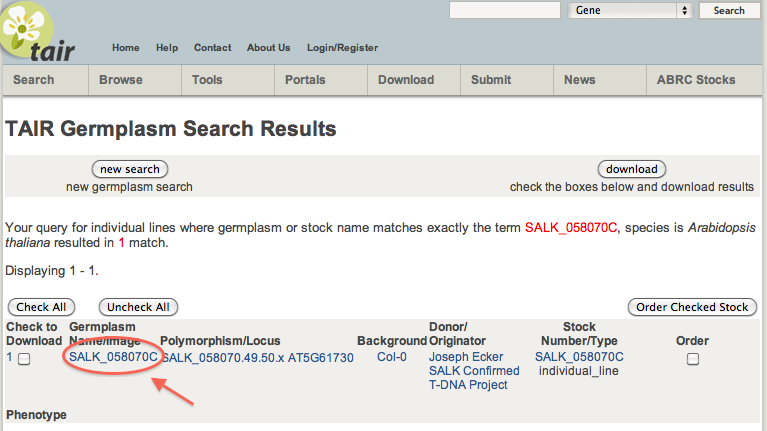
To learn more about your line of interest, the unPAK database is connected to The Arabidopsis Information Resource (TAIR). Clicking the link located above the histograms will connect you to the TAIR search results page for the Arabidopsis line you have selected.
 Copyright secured by Digiprove © 2017
Copyright secured by Digiprove © 2017